Publications
HIGHLIGHTS
2025
* Denotes equal contribution. # Denotes co-corresponding authors. Ferguson Lab members highlighted in bold.
Alkhalaf LM, Arrowsmith C, Balskus EP, Bergamini G, Bhandari R, Chang CJ, Chen P, Chen X, Ciulli A, Cricco JA, Davis BG, Delbianco M, Dudareva N, Dueber E, Ferguson FM, de Giuseppe PO, Hamachi I, Hammond MC, Hatzios SK, Do Heo W, Janet JP, Kamat SS, Knapp S, Krishnan Y, Lang K, Laraia L, Leveson-Gower RB, Li XD, Liu DR, Liu MF, London N, Mahanta N, Mayor-Ruiz C, Muir T, Murakami MT, Rhee HW, Robers M, Satz A, Schulman BA, Shen B, Shoichet B, Strauss E, Suzuki T, Tiwary P, Waldmann H, Ward TR, Weeks A, Weerapana E, Winter G. Thoughts for the future. Nat Chem Biol. 2025;21(1):6-15. doi: 10.1038/s41589-024-01802-2.
2024
Fan AT, Gadbois GE, Huang HT, Chaudhry C, Jiang J, Sigua LH, Smith ER, Wu S, Poirier GJ, Dunne-Dombrink K, Goyal P, Tao AJ, Sellers WR, Fischer ES, Donovan KA, Ferguson FM. A Kinetic Scout Approach Accelerates Targeted Protein Degrader Development. Angew Chem Int Ed Engl. 2024:e202417272. doi: 10.1002/anie.202417272.
Cacace A, Daniels D, Ferguson FM, Mayor-Ruiz C, Woo C. Application of induced proximity for therapeutic discovery. Cell Chem Biol. 2024;31(6):1036-8. doi: 10.1016/j.chembiol.2024.05.015.
Afshar-Sterle S, Carli ALE, O'Keefe R, Tse J, Fischer S, Azimpour AI, Baloyan D, Elias L, Thilakasiri P, Patel O, Ferguson FM, Eissmann MF, Chand AL, Gray NS, Busuttil R, Boussioutas A, Lucet IS, Ernst M, Buchert M. DCLK1 induces a pro-tumorigenic phenotype to drive gastric cancer progression. Sci Signal. 2024;17(854):eabq4888. doi: 10.1126/scisignal.abq4888.
Wang Y, Mouli M, Ma M, Ferguson FM. Making transient complexes stick. Nat Chem Biol. 2024;20(12):1557-8. doi: 10.1038/s41589-024-01649-7.
Tran NL*, Jiang J*, Ma M, Gadbois GE, Gulay KCM, Verano A, Zhou H, Huang CT, Scott DA, Bang AG, Tiriac H, Lowy AM, Wang ES#, Ferguson FM#. ZBTB11 Depletion Targets Metabolic Vulnerabilities in K-Ras Inhibitor Resistant PDAC. bioRxiv. 2024. doi: 10.1101/2024.05.19.594824.
Hangauer MJ, Gutkind JS, Ferguson FM. Pan-RAF:MEK Molecular Glues Take Center Stage. Cancer Discov. doi: 10.1158/2159-8290.CD-24-0539.
Goyal P, Tao AJ, Mumby EJ, English JG#, Ferguson FM#. Measuring Ligand-bound Protein Complexes with Proximity Labeling: A Practical Guide. Chembiochem. 2024;25(10):e202400073. doi: 10.1002/cbic.202400073.
2023
Tao AJ*, Jiang J*, Gadbois GE, Goyal P, Boyle BT, Mumby EJ, Myers SA, English JG#, Ferguson FM#. A biotin targeting chimera (BioTAC) system to map small molecule interactomes in situ. Nature Communications. 2023;14(1):8016. doi: 10.1038/s41467-023-43507-5.
A.T. Fan, B.T. Boyle, F.M. Ferguson. A new avenue for molecular glues: Rapid discovery of a NFKB1 degrader. Cell Chem. Biol., 2023. doi: 10.1016/j.chembiol.2023.04.002
Ferguson FM. PROTACs reach clinical development in inflammatory skin disease. Nat Med. 2023;29(12):3006-7. doi: 10.1038/s41591-023-02622-y.
2022
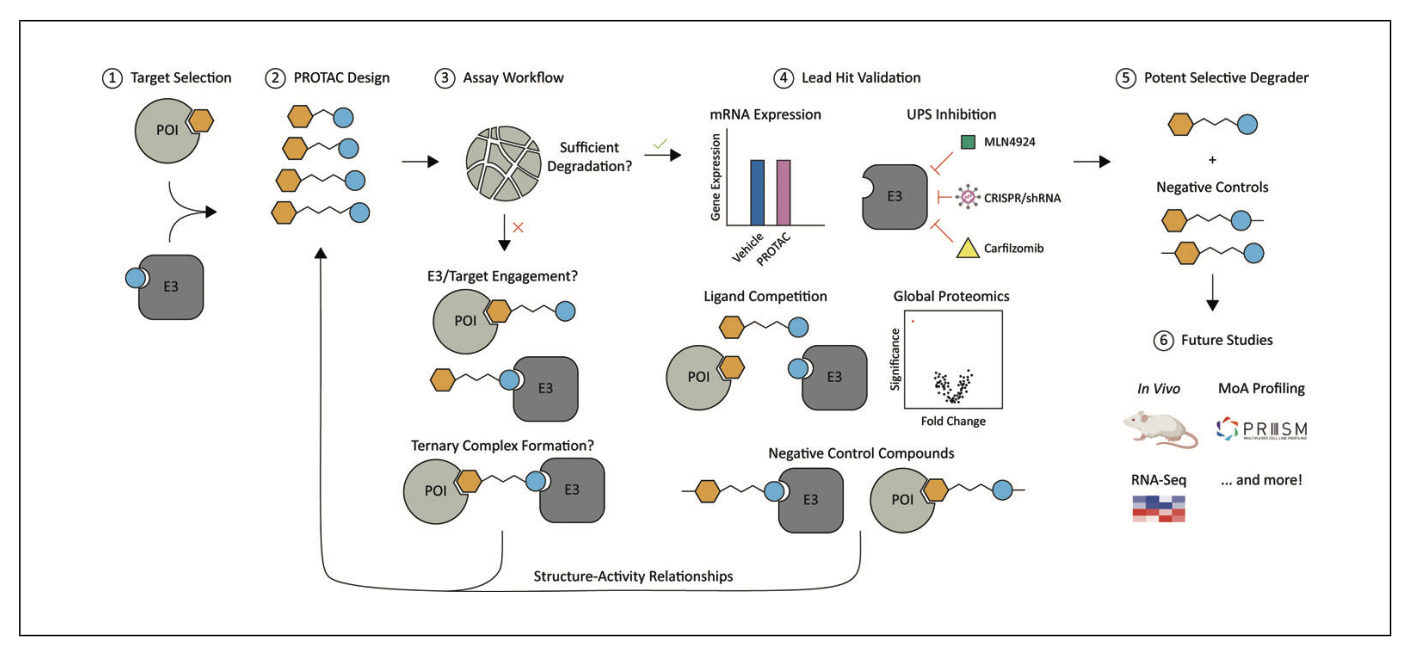
N.L. Tran, G.A. Leconte, F.M. Ferguson. Targeted Protein Degradation: Study Design Considerations for PROTAC Development. Curr. Prot. Mol. Biol. 2022, (in press). doi:10.1002/cpz1.611
Licchesi JDF, Laman H, Ikeda F, Ferguson FM, Bolanos-Garcia VM (2022) Editorial: E3 ubiquitin ligases: From structure to physiology to therapeutics, Volume II. Front. Physiol. 13:1038793. doi: 10.3389/fphys.2022.1038793
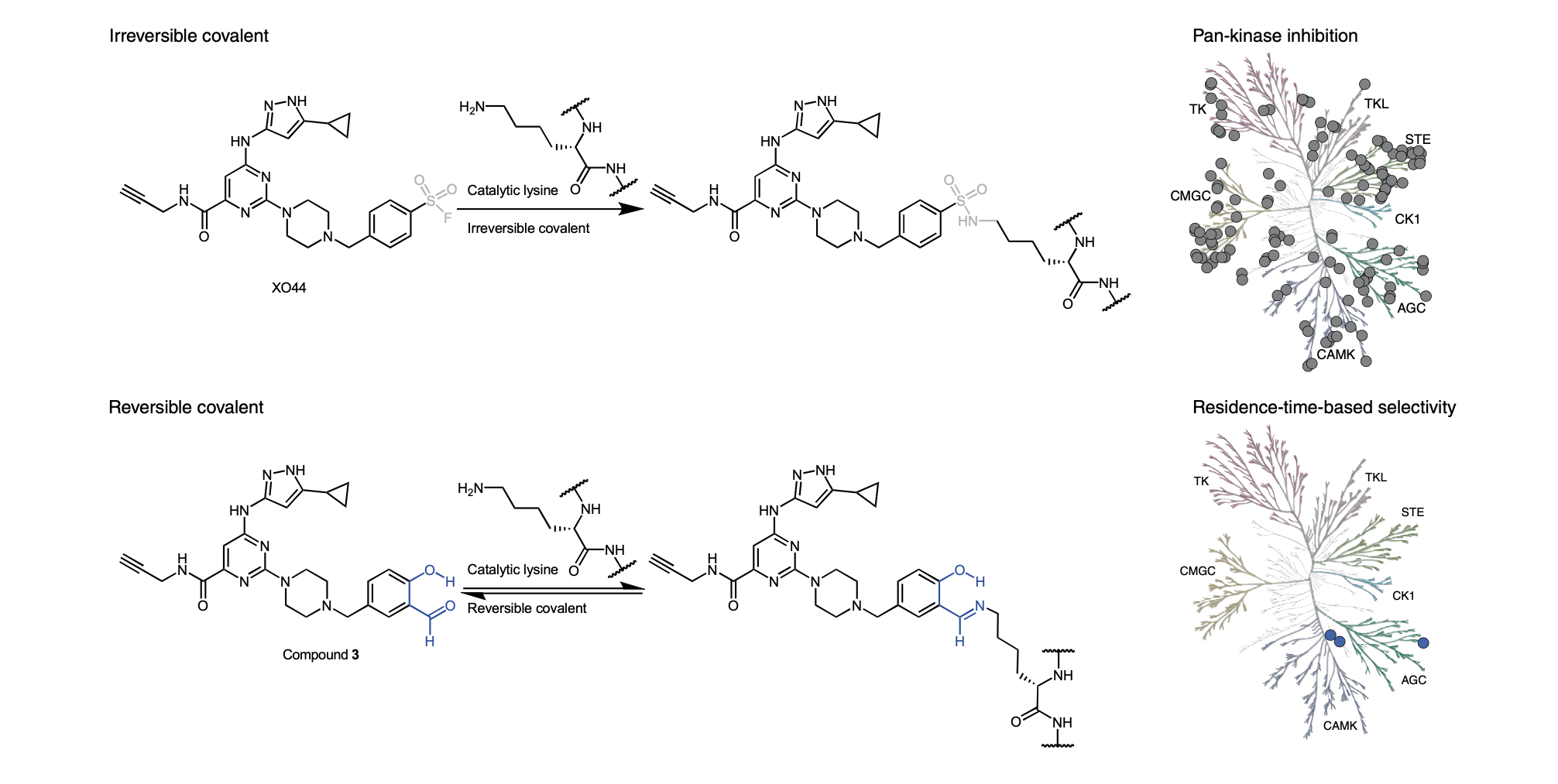
F.M. Ferguson. Tuned Out. Nat. Chem. Biol., 2022, DOI: 10.1038/s41589-022-01037-z
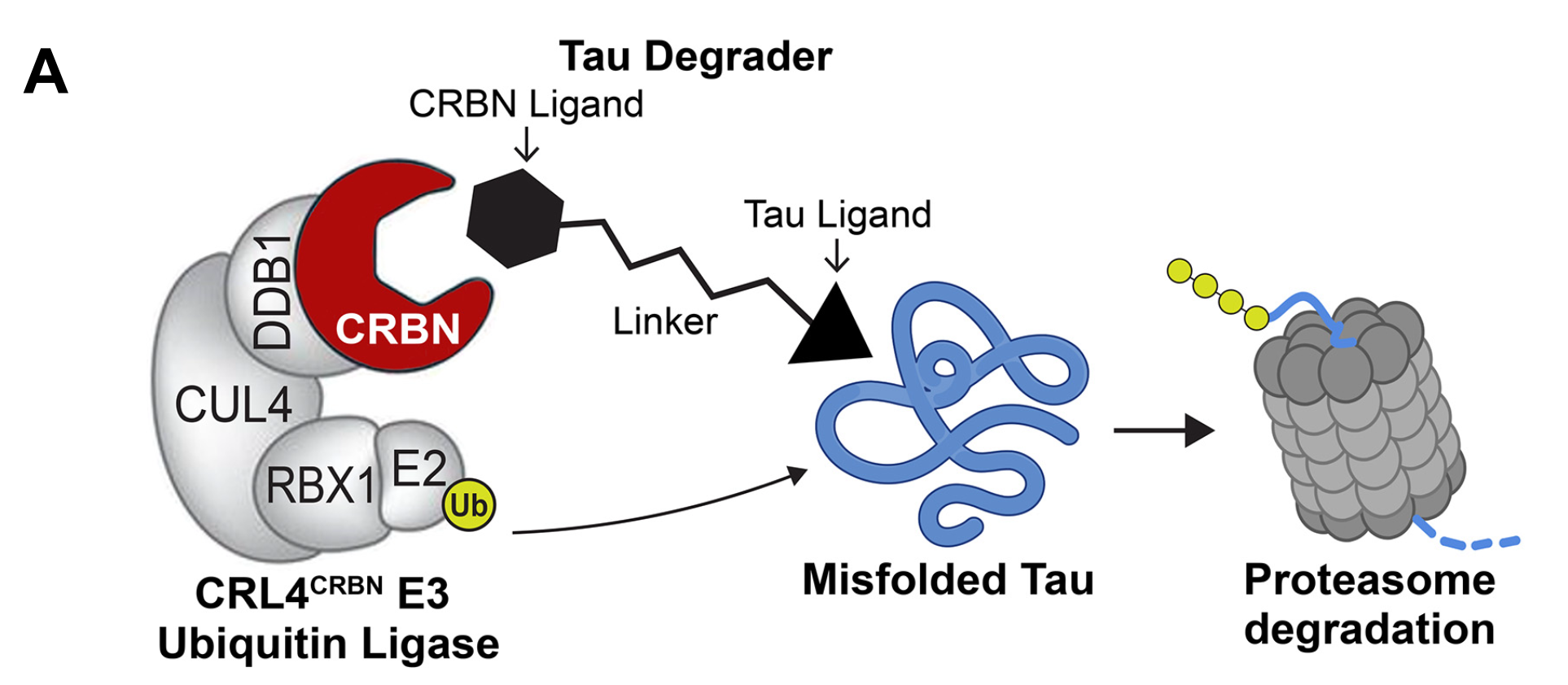
M. C. Silva, G. Nandi, K. A. Donovan, Q. Cai, B. Berry, R. P. Nowak, E. S. Fischer, N. S. Gray, F. M. Ferguson#, S. J. Haggarty#. Discovery and optimization of aberrant tau targeted protein degraders enabled by patient iPSC-derived neuronal tauopathy models. Frontiers in Cellular Neuroscience 2022, DOI: doi.org/10.3389/fncel.2022.801179
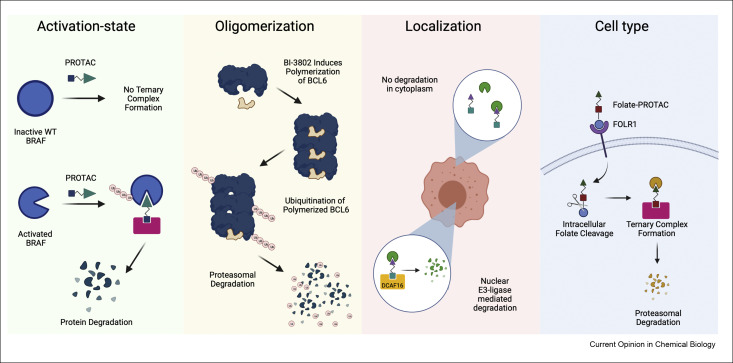
A.J. Tao*, G.E. Gadbois*, S.A. Buczynski*, F.M. Ferguson. Targeted protein degradation: Emerging concepts and protein state-specific targeting principles. Current Opinion in Chemical Biology, 2022, DOI: 10.1016/j.cbpa.2021.102114.
2021
B. K. A. Seong, N. Dharia, S. Lin, K. Donovan, A. Robichaud, A. Conway, A. Hamze, L. Ross, B. Nabet, F. M. Ferguson, B. Stolte, E. Wang, J. Sun, F. Piccioni, N. S. Gray, E. S. Fischer, K. Stegmaier. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma, Cancer Cell, 2021, DOI: 10.1016/j.ccell.2021.07.003.
S. Ryu*, G. E. Gadbois*, A. J. Tao*, B. J. Fram, J. Jiang, B. T. Boyle, K. A. Donovan, N. M. Krupnick, B. C. Berry, D. Bhunia, I. Shin, E. S. Fischer, N. S. Gray#, T. Sim#, F. M. Ferguson#. Synthesis and structure-activity relationships of targeted protein degraders for the understudied kinase NEK9. Current Research in Chemical Biology, 2021, DOI: 10.1016/j.crchbi.2021.100008.
A. L. E. Carli, A. Rai, S. Asfahar-Sterle, H. Fang, R. O’Keefe, J. Tse, F. M. Ferguson, N. S. Gray, M. Ernst, D. Greening#, M. Buchert#. Cancer stem cell marker DCLK1 reprograms small extracellular vesicles toward migratory phenotype in gastric cancer cells. Proteomics, 2021, DOI: 10.1002/pmic.202000098.
B. Jiang*, J. Jiang*, I. H. Kaltheuner*, A. Balboni Iniguez, K. Anand, F. M. Ferguson, S. B. Ficarro, B. K. A. Seong, A. Katrin Greifenberg, S. Dust, N. P. Kwiatkowski, J. A. Marto, K. Stegmaier, T. Zhang#, M. Geyer#, N. S. Gray#. Structure-Activity Relationship Study of THZ531 Derivatives Enables the Discovery of BSJ-01-175 as a Dual CDK12/13 Covalent Inhibitor with Efficacy in Ewing Sarcoma. Eur. J. Med. Chem., 2021, DOI: 10.1016/j.ejmech.2021.113481.
A. J. Tao, F. M. Ferguson. Assembling a robust workflow for characterizing endogenous E3-ligase substrates. Biochemistry, 2021, DOI: 10.1021/acs.biochem.1c00273.
F. M. Ferguson. Harnessing antibody-mimetic selectivity for activation state-specific targeted degradation of endogenous K-Ras. ACS Central Science, 2021, DOI: 10.1021/acscentsci.1c00084.
2013-2020: Graduate and Postdoctoral
* Denotes equal contribution. # Denotes co-corresponding authors.
K. A. Donovan*, F. M. Ferguson*, J. W. Bushman, N. A. Eleuteri, D. Bhunia, S-S Ryu, L. Tan, K. Shi, D. Dobrovolsky, B. Jiang, J. Wang, M. Hao, I. You, M. Teng, Y. Liang, J. Hatcher, Z. Li, T. D. Manz, B. Groendyke, W. Hu, Y. Nam, S. Sengupta, H. Cho, I. Shin, J. Che, S. Buhrlage, T.B. Sim#, N. S. Gray#, E. S. Fischer#. Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell, 2020, DOI: j.cell.2020.10.038.
Y. Liu, F. M. Ferguson, L. Li, M. Kuljanin, W. Harshbarger, S. Gondi, J. Wang, J. D. Mancias, N. S. Gray, K. D. Westover. Chemical biology toolkit for DCLK1 reveals connection to RNA processing. Cell Chem. Biol., 2020, DOI: 10.1016/j.chembiol.2020.07.011.
B. Nabet*, F. M. Ferguson*, B. K. A. Seong, M. Kuljanin, D. L. Buckley, A. L. Leggett, M. L. Mohardt, S. P. Gygi, J. D. Mancias, J. E. Bradner, K. Stegmaier, N. S. Gray. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Comms., 2020, 10.1038/s41467-020-18377-w.
F. M. Ferguson, Y. Liu, W. Harshbarger, L. Huang, J. Wang, S. J. Capuzzi, E. N. Muratov, A. Tropsha, X. Deng, S. Muthuswamy, K. D. Westover, N. S. Gray. Synthesis and structure activity relationships of DCLK1 kinase inhibitors based on a 5,11-dihydro-6H-benzo[e]pyrimido[5,4-b][1,4]diazepin-6-one scaffold. J. Med. Chem., 2020, DOI: 10.1021/acs.jmedchem.0c00596.
M. G. Jaeger, B. Schwalb, S. D. Mackowiak, T. Velychko, A. Hanzl, H. Imrichova, M. Brand, B. Agerer, S. Chorn, B. Nabet, F. M. Ferguson, A. C. Müller, A. Bergthaler, N. S. Gray, J. E. Bradner, C. Bock, D. Hnisz, P. Cramer#, G. E. Winter#. Mechanistic basis for selective Mediator-dependence of cell type-specifying transcription. Nat. Genetics, 2020, DOI: 10.1038/s41588-020-0635-0.
T. D. Manz*, S. C. Sivakumaren*, F. M. Ferguson, T. Zhang, A. Yasgar, M. D. Hall, M. I. Davis, H-S Seo, S. B. Ficarro, J. D. Card, H. Shim, A. T. Sasaki, M. D. Boxer, A. Simeonov, M. Shen, J. A. Marto, S. Dhe-Paganon, L. C. Cantley, N. S. Gray. Discovery and Structure-Activity Relationship Study of (Z)-5-Methylenethiazolidin-4-one Derivatives as Potent and Selective Pan-phosphatidylinositol 5-Phosphate 4-Kinase Inhibitors. J. Med. Chem., 2020, DOI: 10.1021/acs.jmedchem.0c00227.
F. M. Ferguson*, B. Nabet*, S. Raghavan, Y. Liu, A. L. Leggett, M. Kuljanin, R. L. Kalekar, A. Yang, S. He, Ji. Wang, R. W. S. Ng, R. Sulahian, L. Li, E. J. Poulin, L. Huang, J. Koren, N. Dieguez-Martinez, S. Espinosa, Z. Zeng, C. R. Corona, J. D. Vasta, R. Ohi, T. Sim, N. D. Kim, W. Harshbarger, J. M. Lizcano, M. B. Robers, S. Muthaswamy, C. Y. Lin, A. T. Look, K. M. Haigis, J. D. Mancias, B. M. Wolpin, A. J. Aguirre, W. C. Hahn, K. D. Westover, N. S. Gray. Discovery of a selective inhibitor of Doublecortin Like Kinase 1. Nat. Chem. Biol., 2020, DOI: 10.1038/s41589-020-0506-0.
S. C. Sivakumaren*, H. Shim*, T. Zhang*, F. M. Ferguson, M. R. Lundquist, C. M. Browne, B. Jiang, M.-F. Hao, N. P. Kwiatkowski, S. B. Ficarro, M. N. Paddock, D. G. Wang, H.-S. Seo, T. D. Manz, T. J. Yang, P. Krishnan, J. M. Cunningham, S. Dhe-Paganon, J. A. Marto, L. C. Cantley, N. S. Gray. Targeting the PI5P4K lipid kinase family in cancer using novel covalent inhibitors., Cell Chem. Biol., 2020, DOI: 10.1016/j.chembiol.2020.02.003
T. D. Manz, S. C. Sivakumaren, S, A. Yasgar, Dhe-Paganon, M. D. Hall, M. I . Davis, H-S Seo, J. D. Card, S. B. Ficarro, H. Shim, J. A. Marto, S. Dhe-Paganon, A. T. Sasaki, M. B. Boxer, A. Simeonov, L. C. Cantley, M. Shen, T. Zhang#, F. M. Ferguson#, N. S. Gray#. Structure-Activity Relationship Study of Covalent Pan-Phosphatidylinositol 5-Phosphate 4-Kinase Inhibitors. ACS Med. Chem. Lett., 2019, DOI: 10.1021/acsmedchemlett.9b00402.
D. Remillard, D. L. Buckley, H.-S. Seo, F. M. Ferguson, S. Dhe-Paganon, J. E. Bradner, N. S. Gray. Dual inhibition of TAF1 and BRD4 Bromodomains from the BI-2536 Kinase Inhibitor Scaffold. ACS Med. Chem. Lett., 2019, DOI: 10.1021/acsmedchemlett.9b00243.
F. M. Ferguson*, Z. M. Doctor*, S. Ficaro, N. D. Kim, T. Sim, N. S. Gray. Synthesis and structure activity analysis of a series of 4-amino-1H-pyrazoles as covalent inhibitors of CDK14. Bioorg. Med. Chem. Lett. 2019, DOI: 10.1016/j.bmcl.2019.05.024.
M. C. Silva*, F. M. Ferguson*, Q. Cai, K. A. Donovan, G. Nandi, D. Patnaik, T. Zhang, H.-T. Huang, D. E. Lucente, B. C. Dickerson, T. J. Mitchison, E. S. Fischer, N. S. Gray, S. J. Haggarty. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. eLife, 2019, DOI: 10.7554/eLife.45457.
F. M. Ferguson*, Z. M. Doctor*, S. B. Ficaro, C. M. Browne, J. A. Marto, J. L. Johnson, T. M. Yaron, L. C. Cantley, N. D. Kim, T. Sim, M. J. Berberich, M. Kalocsay, P. K. Sorger, N. S. Gray. Discovery of covalent CDK14 inhibitors with pan-TAIRE family specificity., Cell Chem. Biol., 2019, DOI: 10.1016/j.chembiol.2019.02.015.
J. Wang, T. Erazo, F. M. Ferguson, D. L. Buckley, J. Zhang, X. Deng, J. Qi, W. Massefski, J. M. Roberts, P. Cohen, J. Bradner, J. M. Lizcano, S. C. Blacklow, X. Xu, N. S. Gray. Structural and atropisomeric factors governing the selectivity profile of pyrimido-benzodiazipinones as inhibitors of kinases and bromodomains., ACS Chem. Bio., 2018, DOI: 10.1021/acschembio.7b00638.
F. M. Ferguson and N. S. Gray. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov., 2018, DOI:10.1038/nrd.2018.21.
F. M. Ferguson*, Z. M. Doctor*, A. Chikuad, T. Sim, N. D. Kim, S. Knapp, N. S. Gray. Characterization of a highly selective inhibitor of the Aurora kinases., Bioorg. Med. Chem. Lett., 2017, DOI: 10.1016/j.bmcl.2017.08.016.
F. M. Ferguson, J. Ni, T. Zhang, B. Tesar, T. Sim, N. D. Kim, X. Deng, J. R. Brown, J. J. Zhao, N. S. Gray. Discovery of a series of 5,11-dihydro-6H-benzo[e]pyrimido[5,4-b][1,4]diazepin-6-ones as selective PI3K-δ/γ inhibitors., ACS Med. Chem. Lett., 2016, DOI: 10.1021/acsmedchemlett.6b00209.
C. Tallant, E. Valentini, O. Fedorov, L. Overvoorde, F. M. Ferguson, P. Filippakopoulos, D. Svergun, S. Knapp, A. Ciulli. Molecular basis of histone tail recognition by human TIP5 PHD finger and Bromodomain of the chromatin remodelling complex NoRC., Structure, 2015, DOI: 10.1016/j.str.2014.10.017.
M. Baud*, E. Lin Shiao*, T. Cardote*, C. Tallant, A. Pschibul, K-H. Chan, M. Zengerle, J. Rosello, T. Kwan, F. M. Ferguson, A. Ciulli. A Bump-and-Hole Approach to Engineer Controlled Selectivity of BET Bromodomain Chemical Probes., Science, 2014, DOI: 10.1126/science.1249830.
F. M. Ferguson, D. M. Dias, J. P. G. L. M. Rodrigues, H. Weink, R. Boelens, A. M. J. J. Bonvin, C. Abell, A. Ciulli. Binding Hotspots of BAZ2B Bromodomain: Histone Interaction Revealed by Solution NMR Driven Docking, Biochemistry, 2014, DOI: 10.1021/bi500909d.
F. M. Ferguson, O. Fedorov A. Chaikuad, M. Philpott, J. R. C. Muniz, I. Felletar, F. von Delft, T. Heightman, S. Knapp, C. Abell, A. Ciulli. Targeting Low Druggability Bromodomains: Fragment Based Inhibitor Design Against the BAZ2B Bromodomain, J. Med. Chem., 2013, DOI: 10.1021/jm401582c.
