TARGETED PROTEIN DEGRADATION, MOLECULAR GLUES
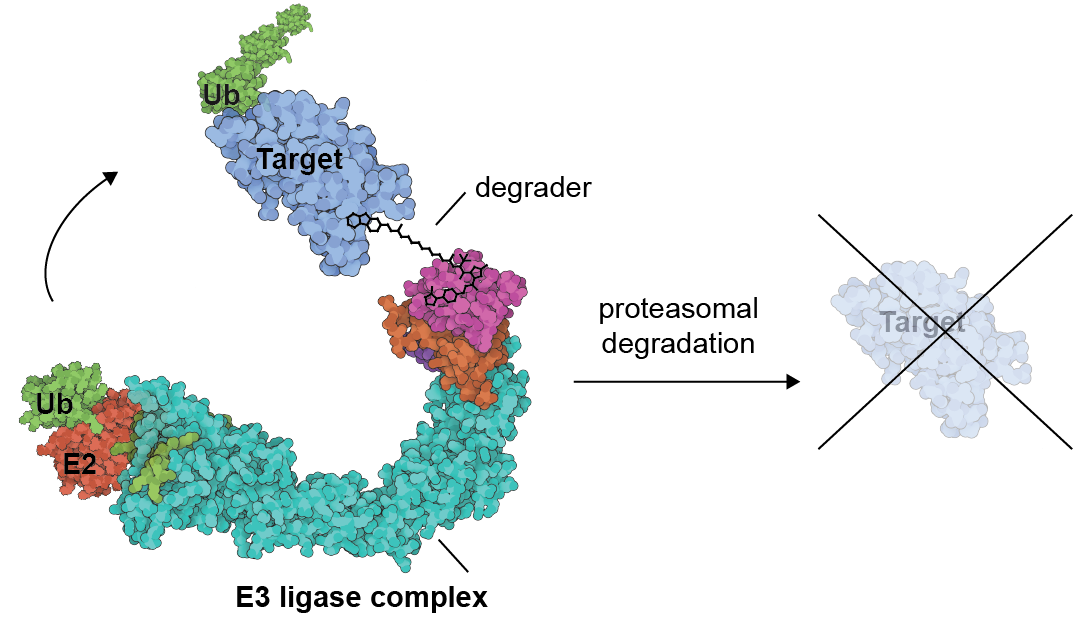
Targeted protein degradation (TPD) is an innovative chemical approach, whereby rather than inhibiting enzymatic function, proteins are selectively targeted for degradation by the proteasome. TPD is of interest in drug development as it can address previously inaccessible targets. Despite this, relatively little is understood about the factors governing a targets propensity for ubiquitination and degradation by specific E3-ligases, and the influence of chemical and cell type specific variables on the effects of a given degrader molecule. We tackle fundamental challenges in degrader design and discovery using chemoproteomic approaches, and develop chemical and chemical genetic degrader tools to answer mechanistic questions about target biology and the ubiquitin proteasome system.
Selected Publications:
Selected Publications:
- A.T. Fan*, G.E. Gadbois*,...F.M. Ferguson. A Kinetic Scout Approach Accelerates Targeted Protein Degrader Development. Angewandte Chemie, 2024, DOI:10.1002/anie.202417272.
- N. L. Tran*, J. Jiang*, M. Ma, G. Gadbois, K. C. M. Gulay, A. Verano, H. Zhou, C.-T. Huang, D. A. Scott, A. G. Bang, H. Tiriac, A. M. Lowy, E. S. Wang#, F.M. Ferguson#. ZBTB11 Depletion Targets Metabolic Vulnerabilities in K-Ras-inhibitor Resistant PDAC. Nature Chemical Biology, 2025, DOI: 10.1038/s41589-025-01978-1
CHEMICAL CONTROL OF MISFOLDED PROTEIN FATE
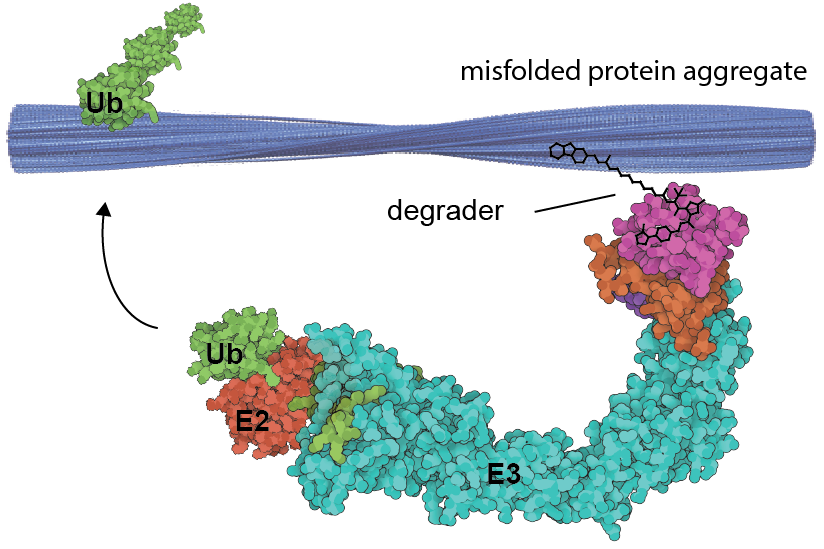
The accumulation of misfolded proteins concurrent with disease progression is a hallmark of neurodegenerative disorders known collectively as proteinopathies. An important class of these is the tauopathies, such as Alzheimer’s disease (AD) and Frontotemporal dementia (FTD), which are linked to the misfolding of tau protein. However, how these aggregated species contribute to disease etiology and progression remains unclear. A key challenge in the field is the lack of tool compounds able to exert spatiotemporal control over misfolded protein levels that can be used to interrogate their function across multiple disease models. We harness TPD and related induced proximity technologies to develop potent degraders for proteinopathies, and study their effects in patient derived models of disease.
Selected Publications:
Selected Publications:
- M. C. Silva*, F. M. Ferguson*, et al. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. eLife, 2019, DOI: 10.7554/eLife.45457.
- M. C. Silva, G. Nandi, K. A. Donovan, Q. Cai, B. Berry, R. P. Nowak, E. S. Fischer, N. S. Gray, F. M. Ferguson#, S. J. Haggarty#. Discovery and optimization of aberrant tau targeted protein degraders enabled by patient iPSC-derived neuronal tauopathy models. Front. Cell. Neurosci. 2022, DOI: 10.3389/fncel.2022.801179
PROXIMITY-MEDIATED PHARMACOLOGY
We develop new tools (heterobifunctional dimerizers and molecular glues) allowing us to investigate the effects of interactome remodeling on protein misfolding, accumulation, and disease phenotypes. In addition to the presence of misfolded proteins, neurodegenerative diseases are driven by misregulated protein postranslational modifications (PTMs), such as phosphorylation, acetylation, and methylation. The enzymes that write and erase these marks have multiple substrates, limiting the precision with which we can manipulate PTMs on a single protein of interest in living cells. We harness proximity pharmacology to develop precision tools for these proteins.
Selected Publications
Selected Publications
- A.J. Tao*, J. Jiang*, ..J.G. English#, F.M. Ferguson#. A biotin targeting chimera (BioTAC) system to map small molecule interactomes in situ. Nature Communications, 2023, DOI: 10.1038/s41467-023-43507-5
- P. Goyal, ...J.G. English#, F.M. Ferguson# Measuring Ligand-bound Protein Complexes with Proximity Labeling: A Practical Guide. ChemBioChem, 2023, DOI: 10.1002/cbic.202400073
M. Ma*, Y. Wang*, J. Koeckenberger, A. J. Tao, E. Mumby, J.Jiang, S. S. V. M. Madaka, J. G. English, F. M. Ferguson. The APEXTAC System for Ligand-guided Proximity Labeling. ChemBioChem, 2025, DOI: 10.1002/cbic.202500602

